MuT-POWER - Mu transposition pooling method with sequencer
Schema of the MuT-POWER method.
Initial 8 fold pooling of the G1 DNA samples with a later 4 fold pooling for the capillary electrophoresis step increases the overall efficiency of this method compared to standard sequencing dramatically.
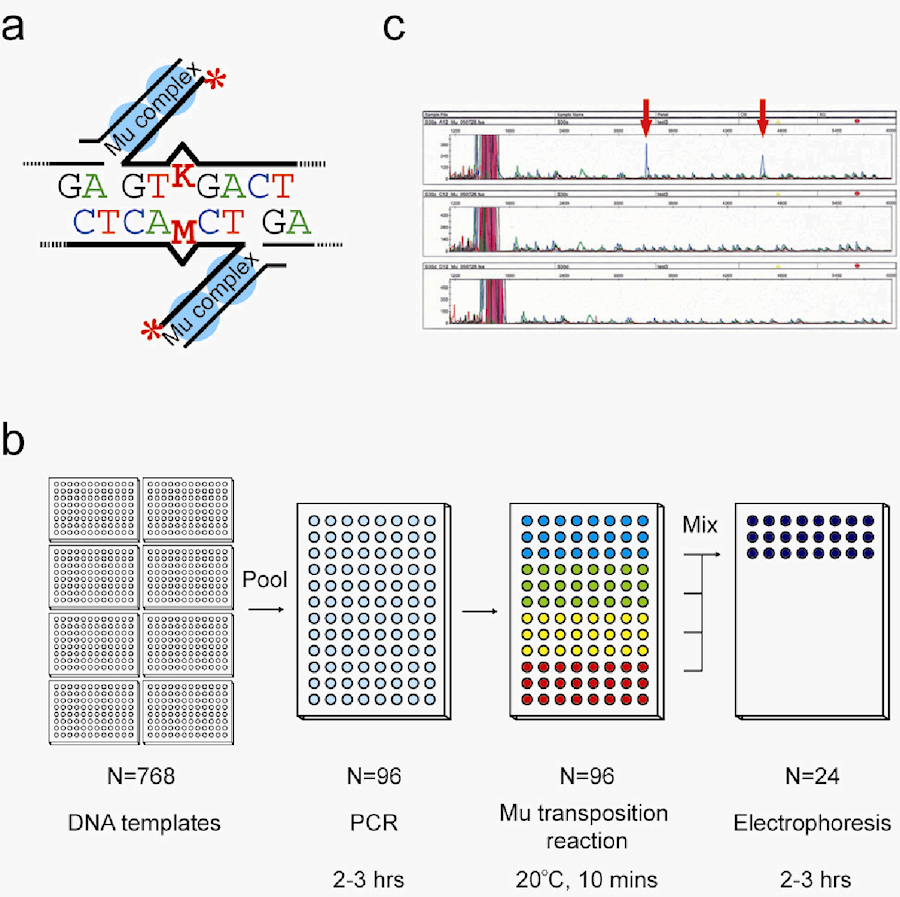
(a) Detection of single-nucleotide mismatches by Mu transposition. The Mu transpososome is a complex of Mu-end DNA and MuA transposase. The Mu-end DNA fragments are labeled at the 5’-ends indicated as red asterisks. The mismatched DNAs are targeted by the Mu transpososome to generate two reaction products indicated as thick lines. (b) Schematic of the high-throughput DNA screening method, MuT-POWER (Mu transposition pooling method with sequencer), for ENU-mutagenized G1 rats. Genomic DNA of G1 rats is extracted and pooled eightfold for PCR. Targeted genomic regions are amplified by PCR for 2-3 hours. PCR products are then mixed with Mu transposase and fluorescence-labeled Mu-end DNA: FAM (blue), HEX (green), NED (yellow) and ROX (red), and incubated at 20°C for 5min. Thereafter, Mu reaction products are mixed as one sample. The reaction mixtures are capillary-electrophoresed by an automatic sequencer. (c) Transposition reaction products are easily visualized as two positive peaks by Gene-Mapper software. The mutations are confirmed by sequencing each DNA from the pooled 8 G1 rats.